Sie befinden sich hier
Inhalt
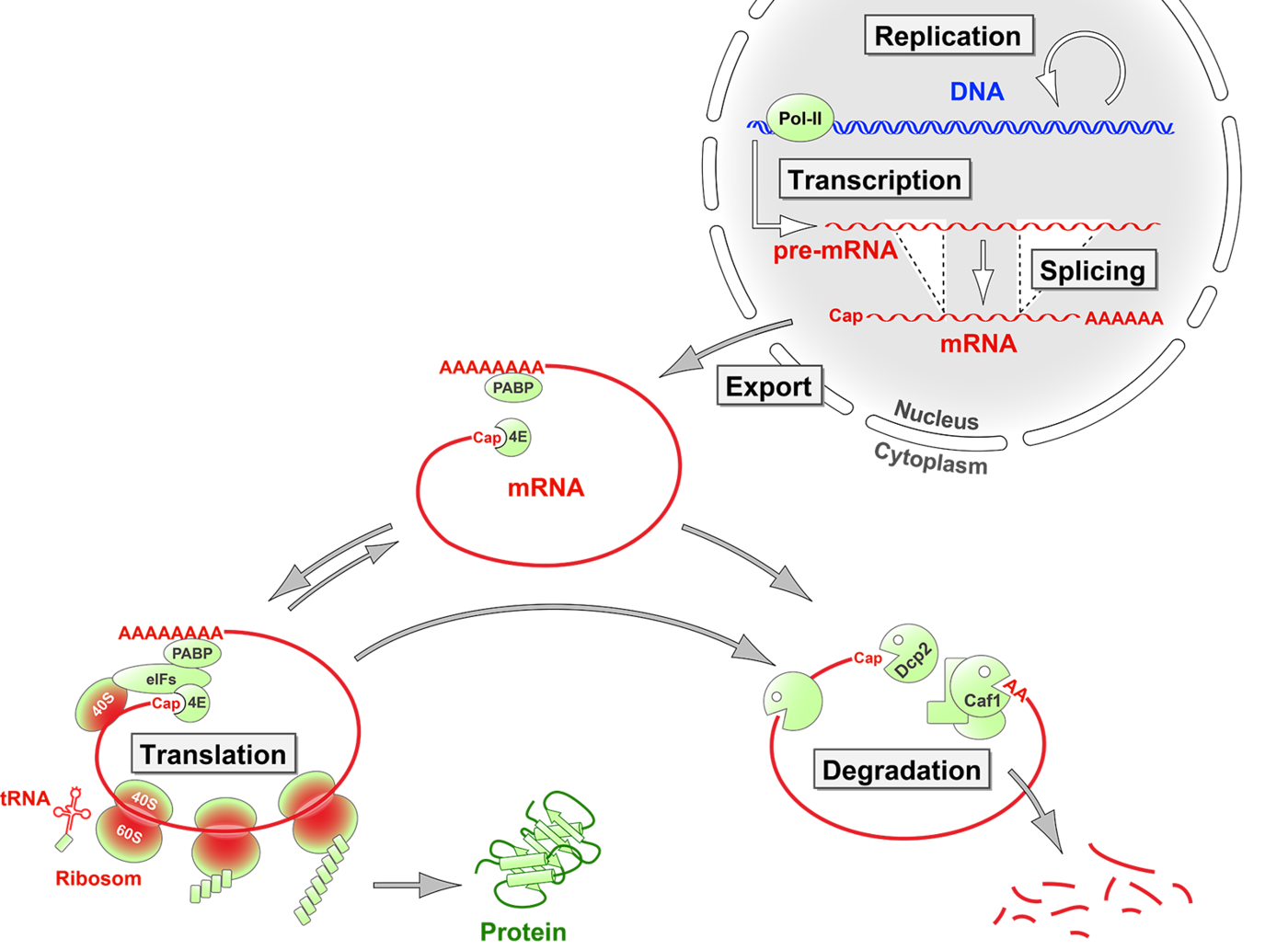
Mechanisms of mRNA turnover and translation control
Once an mRNA is exported from the nucleus, the rate at which it is translated into the encoded polypeptide represents an efficient and rapid means to regulate gene expression at the posttranscriptional level. Likewise, the active control of mRNA degradation rates allows cells to shape the dynamics of gene expression, and rapidly turn off specific genes. We investigate cis-acting RNA elements, trans-acting RNA-binding proteins and signaling pathways that specifically determine the fate of individual mRNAs. In addition, we uncover mechanisms by which cells control the global rates of mRNA turnover and translation. More recently, we have also started to explore the role of RNA-binding proteins in maintenance of genome stability and cell cycle checkpoints. You can find details on some of our discoveries under Highlights.
Current projects in the lab focus on:
- the central role of the Ccr4-Caf1-Not deadenylase complex in controlling mRNA decay
- the impact of mRNA modifications on mRNA translation & turnover in macrophages
- the design of immune-modulatory RNAs
- mechanisms regulating translation elongation
- translation control by RNA viruses
- cellular responses to ribotoxic stress
- RNA-binding proteins during the replication stress response
Selected publications
Poetz F, Corbo J, Levdansky Y, Spiegelhalter A, Lindner D, Magg V, Lebedeva S, Schweiggert J, Schott J, Valkov E, Stoecklin G. RNF219 attenuates global mRNA decay through inhibition of CCR4-NOT complex-mediated deadenylation. Nat Commun 2021; 12:7175.
Schott J, Reitter S, Lindner D, Grosser J, Bruer M, Shenoy A, Geiger T, Mathes A, Dobreva G, Stoecklin G. Nascent Ribo-Seq measures ribosomal loading time and reveals kinetic impact on ribosome density. Nat Methods 2021; 18:1068-1074.
Haneke K, Schott J, Lindner D, Kruse Hollensen A, Kroun Damgaard C, Mongis C, Knop M, Palm W, Ruggieri A, Stoecklin G. CDK1 couples proliferation with protein synthesis. J Cell Biol 2020; 219:e201906147.
Lafarga V, Sung HM, Haneke K, Roessig L, Pauleau AL, Bruer M, Rodriguez-Acebes S, Lopez-Contreras AJ, Gruss OJ, Erhardt S, Mendez J, Fernandez-Capetillo O, Stoecklin G. TIAR marks nuclear G2/M transition granules and restricts CDK1 activity under replication stress. EMBO Rep 2019; 20:e46224.
Sharma S, Poetz F, Bruer M, Ly-Hartig TBN, Schott J, Séraphin B, Stoecklin G. Acetylation-Dependent Control of Global Poly-A RNA Degradation by CBP/p300 and HDAC1/2. Mol Cell 2016; 63:927-38.
Schott J, Reitter S, Philipp J, Haneke K, Schäfer H, Stoecklin G. Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLoS Genet 2014; 10:e1004368.
Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell, 2013; 153:869-81.
Kontextspalte
Head of Department

Prof. Dr. Dr. Georg Stoecklin
Phone +49 621 383-71444
georg.stoecklin@medma.uni-heidelberg.de
