Sie befinden sich hier
Inhalt
Nuclear speckles: sites of stress-induced gene expression and splicing
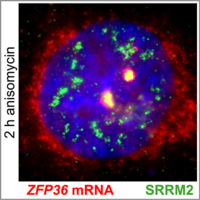
Traditionally, nuclear speckles (NSs) are considered reservoirs of splicing factors that facilitate posttranscriptional mRNA processing. We discovered that ribotoxic stress induces a profound reorganization of NSs with enhanced recruitment of RNA polymerase II and factors required for splice-site recognition. NS reorganization is accompanied by relocalization of immediate early gene transcription foci to nuclear speckles and pronounced splicing activation of the corresponding pre-mRNAs. Our study assigns nuclear speckles a new function as sites dedicated to the preferential expression of stress-induced genes. (Sung et al, 2023).
Queuosine-tRNA promotes sex-dependent learning and memory
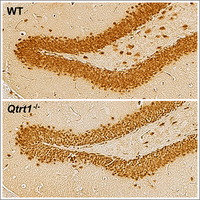
Queuosine (Q) is a modified nucleoside introduced into specific tRNAs by a complex comprising queuine tRNA ribosyltransferase 1 (QTRT1). We discovered that loss of Q-tRNA modification by knockout of the Qtrt1 gene in mice leads to an imbalance in codon-biased protein translation speed, resulting in alterations of hippocampal architecture and sex-dependent reduction in learning and memory formation (Cirzi et al, 2023).
RNA-binding protein CPEB4 represses immediate early gene expression
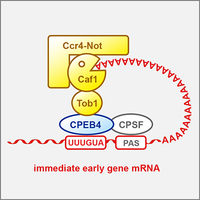
Cytoplasmic polyadenylation element-binding protein 4 (CPEB4) is an RNA-binding protein known to control poly(A) tail length of mRNAs. We show that CPEB4 forms a repressor complex by associating with the Ccr4-Caf1-Not deadenylase machinery, and promotes the degradation of many immediate early gene mRNAs. Our crosslinking results revealed that CPEB4 preferentially binds to G-containing variants of the cytoplasmic polyadenylation element (Poetz et al., 2022).
E3 ligase RNF219 inhibits the Ccr4-Caf1-Not deadenylase complex
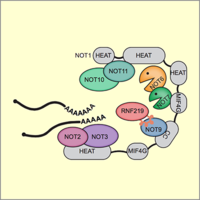
The Ccr4-Caf1-Not complex is a major cellular deadenylase that drives widespread mRNA degradation. Through purification of the endogenous Not1 subunit, we could identify novel interactors of the complex. For one of them, the E3 ubiquitin ligase RNF219, we could demonstrate that it acts as an inhibitor of the Ccr4-Caf1-Not deadenlyase and stabilizes a wide range of mRNAs. Moreover, we could demonstrate that a short linear motif within RNF219 is responsible for its interaction with the Not9 subunit of the complex (Poetz et al., 2021).
Nascent Ribo-Seq measures ribosome loading time
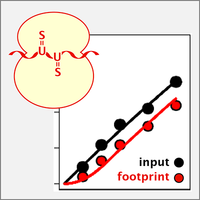
By ribosome profiling (Ribo-Seq), translation of individual mRNAs can be assessed on a transcriptome-wide scale and down to codon resolution. We now developed nascent Ribo-Seq (nRibo-Seq) whereby we could measure - for the first time - the speed by which newly synthesized mRNAs are loaded with ribosomes. Surprisingly, our measurements showed that bulk mRNA requires a considerable amount of time to be fully loaded with ribosomes, between ~40 min in macrophages and ~70 min in mouse embryonic stem cells. Moreover, we discovered that ribosome loading is strongly accelerated on short-lived mRNAs and differs between functional mRNA categories (Schott et al., 2021).
CDK1 enhances global protein synthesis
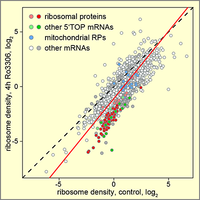
By searching for novel regulators of translation using a stress granule-based screen, we discovered that CDK1 acts as an enhancer of global protein synthesis. Strongest activation was observed for the synthesis of ribosomal proteins, all of which are encoded by 5'TOP mRNAs. Notably, this effect of CDK1 was dependent on the 5'TOP-binding protein LARP1, a known regulator of ribosomal protein synthesis. Our study provides evidence that CDK1 plays a central role in connecting cell proliferation with cell growth. (Haneke et al., 2020).
TIAR maintains G2/M cell cycle checkpoint and ensures genome stability
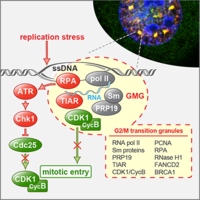
TIAR is an RNA-binding protein that participates in translational suppression and the assembly of cytoplasmic stress granules. We discovered that TIAR has an additional function by maintaining the G2/M cell cycle checkpoint under conditions of replication stress. We could further show that TIAR suppressed the activity of CDK1, a kinase essential for progression through S-phase and entry into mitosis. When cells are arrested at the G2/M boundary, TIAR recruits CDK1 into a newly identified nuclear subdomain we termed G2/M transition granules (GMGs), indicating that GMGs participate in maintenance of the G2/M checkpoint (Lafarga et al., 2019).
Ccr4-Caf1-Not complex acetylation enhances global mRNA decay
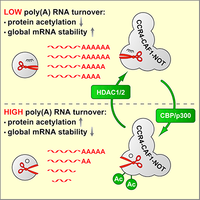
mRNA degradation is initiated by shortening of the poly(A) tail through the Pan2-Pan3 and the Ccr4-Caf1-Not deadenylase complexes. We discovered that class I HDAC inhibitors strongly promote the degradation of global poly(A) RNA, and could identify specific lysine residues within the Caf1 exoribonuclease whose acetylation enhances the deadenlyase activity of the Ccr4-Caf1-Not complex. This mechanism may be important during cellular differentiation, where we observed a global stabilization of mRNA (Sharma et al., 2016).
Control of mRNA translation in macrophages
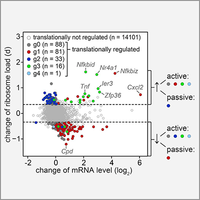
By polysome profiling, we identified mRNAs whose translation is actively regulated in macrophages upon stimulation with bacterial lipopolysaccharide. Since many of the identified mRNAs encode feedback inhibitors of NfkB signaling, translation control is important for downregulating inflammatory responses. In this study we could further demonstrate that translationally regulated Ier3 contributes the survival of activated macrophages (Schott et al., 2014).
A novel class of stem-loop RNA decay elements
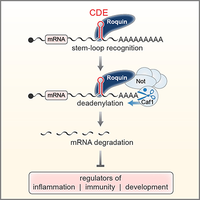
Based on our earlier description of the Constitutive Decay Element (CDE) in the 3'UTR of TNF mRNA (Stoecklin et al., 2003), we determined that the CDE is a stem-loop RNA motif and identified Roquin as a stem-loop specific CDE-binding protein. Our work further showed that Roquin promotes mRNA degradation by recruiting the Ccr4-Caf1-Not deadenylase complex. The CDE represents a novel class of RNA degradation motifs with more than 50 highly conserved CDEs that could be identified in vertebrate mRNAs (Leppek et al., 2013). In collaboration with Teresa Carlomagno (EMBL Heidelberg), the structure of the CDE RNA stem-loop was solved by NMR (Codutti et al., 2015).
AU-rich elements drive rapid degradation of cytokine and IEG mRNAs
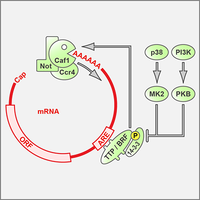
Tristetraprolin (TPP) is an RNA-BP that binds to AU-rich elements (AREs) and causes rapid degradation of its target mRNAs, thereby suppressing the expression of cytokine and immediate early gene (IEG) mRNAs (Schott & Stoecklin, 2010). Our work showed that TTP induces mRNA degradation by recruiting the Ccr4-Caf1-Not complex (Sandler et al., 2011), and that the activity of TTP is inhibited by phosphorylation and 14-3-3 binding (Stoecklin et al., 2004). We improved a method to capture RNA-binding proteins using a streptavidin-binding aptamer, which allowed us to isolate almost all of the currently known ARE-binding proteins (Leppek et al., 2014). Using a forward genetics approach, we earlier identified BRF1 as a paralog of TTP that also promotes the degradation of ARE-containing mRNAs (Stoecklin et al., 2002).
Kontextspalte
Biochemie
TPMA2
Franz-Volhard-Str. 6
68167 Mannheim
Phone +49 621 383-71430
biochemie@medma.uni-heidelberg.de
Head

Prof. Dr. Dr. Georg Stoecklin
Phone +49 621 383-71444
georg.stoecklin@medma.uni-heidelberg.de