Sie befinden sich hier
Inhalt
Translational control by tRNA modifications
Of the more than 170 distinct RNA modifications, more than 90 are found in the 40 known tRNAs. These modifications are located on specific residues including the tRNA anticodon loop, and vary among the three kingdoms of life. Modified nucleotides influence all aspects of tRNA activity and are deposited at different steps during tRNA maturation. We aim to elucidate the biological function of these modifications with a special focus on 5-methylcytidine and queuosine. In particular, we study the molecular mechanisms that couple tRNA modifications to tRNA processing, with a focus on tRNA splicing, and examine how tRNA modifications contribute to the efficiency and accuracy of protein synthesis. We further explore the importance of tRNA modifications for the physiology of multicellular organisms using two organismal models, Caenorhabditis elegans and Mus musculus.
Figure legend:
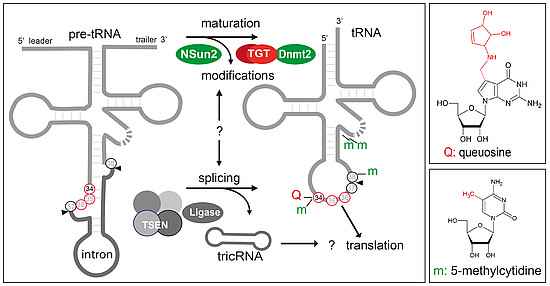
In eukaryotes, methylation of carbon 5 in cytidine (5-methylcytidine) it is catalysed by Nsun2 at positions 48, 49, 50 of the majority of tRNAs, and in the anticodon loop at positions 34 of tRNALeu. A second enzyme, Dnmt2, methylates position 38 of tRNAAsp, tRNAGly and tRNAVal. Queuosine (Q) is a hyper-modified guanosine analogue that comprises a 7-deaza-guanine core structure covalently linked to an amino-methyl side chain and a cyclo-pentanediol moiety. The tRNA guanine transglycosylase, TGT, which comprises the Qtrt1 and Qtrt2 subunits, introduces Q at position 34 of tRNAAsp, tRNAAsn, tRNATyr and tRNAHis. tRNAs are transcribed as precursor molecules that undergo several maturation steps. For certain tRNAs like tRNALeu, tRNAIle and tRNATyr, processing involves excision of an intron located between nucleotides 37 and 38 of the mature tRNA, adjacent to the anticodon loop (positions 34 to 36). This non-conventional splicing mechanism requires separate enzyme-catalysed cleavage (TSEN) and ligation (Ligase) reactions. Site-specific processing and modification of tRNAs are tightly controlled and interdependent processes.
Selected publications
Cirzi C, Dyckow J, Legrand C, Schott J, Guo W, Perez Hernandez D, Hisaoka M, Parlato R, Pitzer C, van der Hoeven F, Dittmar G, Helm M, Stoecklin G, Schirmer L, Lyko F, Tuorto F. Queuosine-tRNA promotes sex-dependent learning and memory formation by maintaining codon-biased translation elongation speed. EMBO J. 2023 Aug 23:e112507.
Navarro IC, Tuorto F, Jordan D, Legrand C, Price J, Braukmann F, Hendrick A, Akay A, Kotter A, Helm M, Lyko F, Miska EA. Translational adaptation to heat stress is mediated by 5-methylcytosine RNA modification in Caenorhabditis elegans. (2020). EMBO J. e105496.
Legrand C, Tuorto F. (2020). RiboVIEW: a computational framework for visualization, quality control and statistical analysis of ribosome profiling data. Nucleic Acids Research. 48, e7.
Tuorto F, Parlato R. (2019). rRNA and tRNA Bridges to Neuronal Homeostasis in Health and Disease. J Mol Biol. 19: 30122-6. Review.
Tuorto F, Legrand C, Cirzi C, Federico G, Liebers R, Müller M, Ehrenhofer-Murray AE, Dittmar G, Gröne HJ, Lyko F. (2018). Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. e99777.
Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M, Lyko F. (2017). Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 27: 1589-1596.
Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Gröne HJ, Dittmar G, Glimm H, Lyko F. (2015). The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 34:2350-2362.
Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. (2012). RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 19:900-5.
Kontextspalte
Project Group Leader

Dr. rer. nat. Francesca Tuorto
Phone +49 621 383-71439
francesca.tuorto@medma.uni-heidelberg.de