Sie befinden sich hier
Inhalt
The NGS core facility is running two different NGS technologies, each of which have advantages/disadvantages depending on the application. Specifically, four different NGS platforms including the Illumina NovaSeq6000, Illumina NextSeq550 as well as an IonTorrent PGM System are being used. In order to allow single cell sequencing, the lab runs a 10x Genomics platform that uses droplet technology to generate single cell suspensions. All platforms are run as operator-based systems.
Illumina NovaSeq6000
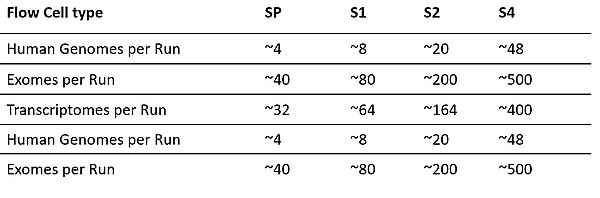
The NovaSeq6000 system performs large-scale sequencing efficiently and cost-effectively. Its tunable output generates up to 6 Tb and 20 B single reads in dual flow cell mode with streamlined workflows. The system offers sequencing of up to 48 genomes, 500 exomes or 400 transcriptomes in ~2 days in a single run with comprehensive coverage.
Features
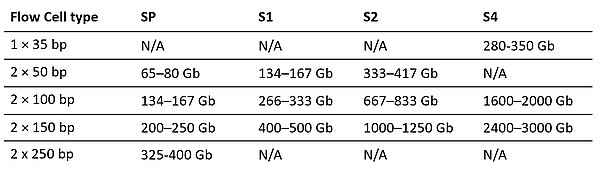
- Scalability (80 - 6000 Gb) in a single run supporting a broad range of applications and study sizes
- Run time: 13–44 hours
- Proven SBS chemistry with single-base extension enables accurate sequencing of homopolymers
- Patterned flow cell technology to generate an unprecedented level of throughput for a broad range of sequencing applications
- Fully automated paired-end sequencing
- Mix and match flow cell types, and run one or two flow cells at a time
- 4 Flow Cell Type (SP, S1, S2, S4)
- Max read length: 2 × 250 bp
- From 800 million reads per end per flow cell (SP) up to 10 Billion reads per end per flow cell (S4)
- Preadjustment of library diversity is required for optimum sequence quality
Data Generation
Reads Passing Filter Per Flow Cell

Sequencing flexibility
- Output per Flow Cell for Various Read Lengths
- Set-up options include single-read or paired-end runs
Instrumentation
Laser
Wavelengths: 532 nm, 660 nm, 780 nm, 790 nm
Instrument Control Computer
- Memory: 2 × 8 GB DDR3L SODIMM
- Base Unit: Portwell WADE-8022 with Intel i7 4700EQ CPU
- Solid-State Drive: 256 GB mSATA
- Operating System: Windows 10
Sequencing Run Time

Illumina NextSeq 550

The Illumina NextSeq® 550 is the largest desktop next-generation sequencing (NGS) system from Illumina capable of sequencing a 30x human genome in a single run. Two different flow cell options and multiple reagent configurations are providing a maximum application flexibility (130 to 400M clusters; 40 to 120 Gb).
Accordingly, the system is suitable for a high number of different applications, e.g. whole exome- or genome sequencing, RNAseq, targeted resequencing or gene expression profiling and single-cell sequencing. The sequencing technology is based surface-based bridge amplification for clonal amplification and sequencing of each generated cluster using the Illumina "sequencing by synthesis" (SBS) method with reversible terminator chemistry.
Features
- Scalability (20–120 Gb) in a single run supporting a broad range of applications and study sizes
- Sequencing runs, including on-board cluster generation, complete in 12–30 hours
- Proven SBS chemistry with single-base extension enables accurate sequencing of homopolymers (sometimes a problem on the IonTorrent system)
- Fully automated paired-end sequencing
- Automated BeadChip array scanning and image file generation
- Approximately 7000 peer-reviewed publications have been published using Illumina SBS sequence data, and more than 24,000 peer-reviewed publications have been published using Illumina array technology
- Number of reads per run
- preadjustment of library diversity is required for optimum sequence quality
Data Generation
Throughput per run
- Up to 400 million reads per run (eg, clusters passing filter) using High Output flow cell
- Up to 130 million reads per run (eg, clusters passing filter) using Mid Output flow cell
Sequencing flexibility
- 100–120 Gb data per 2 × 150 bp run using High Output flow cell and reagents
- 33–40 Gb data per 2 × 150 bp run using Mid Output flow cell and reagents
- Set-up options include single-read or paired-end runs
Array Scanning
- Flow cell options (eg, Mid or High Output flow cells) can be used to select data output levels
- Read length is fully adjustable up to 300 base pairs
- Single array imaged per scanning session
- Up to 12 samples imaged per run with current array support
Instrumentation
Illumination
- 12 light-emitting diodes at 520 nm, 650 nm
Instrument Control Computer
- Dual Intel Xeon E5-2448L 1.8 GHz CPU with 96 GB of RAM included for instrument control, processing images, and base calling
- Conducts real-time analysis processing that automatically produces image intensities and quality-scored base calls directly on the instrument computer
Sequencing run time
- ~ 12 hours for a 1 × 75 bp single-read sequencing run
- ~ 18 hours for a 2 × 75 bp paired-read sequencing run
- ~ 30 hours for a 2 × 150 bp paired-read sequencing run
Ion Torrent PGM

The DNA-Sequencing on the desktop NGS system PGM from Ion Torrent Life Technologies is based on the principle of Semiconductor Sequencing developed in 2007. This technology allows a real-time measurement of hydrogen ions produced during DNA replication and thereby directly translates genetic information (DNA) to digital information (DNA sequence).
This is enabled by an ion semiconductor chip on which the sequencing reaction takes place. The different available chips allow a flexible output for ultimate project flexibility. Additonally, the sequencing times are short, e.g. 30 min for a 1 x 35 bp on a 314 chip or 8 hours for a 1 x 400 bp run on a 318 chip.
Data Generation
Number of reads per run
- Up to 0.5 million reads per Ion 314 chip (eg, reads passing filter)
- Up to 3 million reads per Ion 316 chip (eg, reads passing filter)
- Up to 5.5 million reads per Ion 318 chip (eg, reads passing filter)
Throughput per run
- 30-50 Mb per 1 x 200 bp reads and 60-100 Mb per 1 x 400bp read on a Ion 314 Chip
- 300-600 Mb per 1 x 200 bp reads and 600 Mb - 1 Gb per 1 x 400bp read on a Ion 316 Chip
- 600 Mb - 1 Gb per 1 x 200 bp reads and 1.2 - 2 Gb per 1 x 400bp read on a Ion 318 Chip
Instrumentation
Configuration
A single free-standing tower computer appliance, included with the purchase of the Ion PGM™ System. Includes Torrent Suite™ Software with all necessary software components to deliver signal processing, base calling, read alignment, and variant calling.
Instrument Control Computer
- Dual 6-core 2.66 GHz CPUs, 48 GB RAM, 16 TB Storage
Sequencing run time
- ~ 2 hours per 1 x 200 bp reads and ~ 4 hours per 1 x 400bp read on a Ion 314 Chip
- ~ 3 hours per 1 x 200 bp reads and ~ 5 hours per 1 x 400bp read on a Ion 316 Chip
- ~ 4.5 hours per 1 x 200 bp reads and ~ 7 hours per 1 x 400bp read on a Ion 318 Chip
10x Genomics Chromium iX

The Chromium iX allows for high-throughput single cell transcriptome, immunome, multiome or cell surface protein analyses in a variety of cell types as wells as nuclei from fresh or fixed samples (including FFPE). The flexible workflow encapsulates single cells or nuclei together with gel beads intro nanodroplets. The underlying GEMCode Technology enables the execution of thousands of micro-reactions in parallel. A GEM is a „GelBead in EMulsion“ droplet that encapsulates each micro-reaction within the Chromium System. 10x Genomics offers a multitude of kit solutions including gene expression, chromatin accessibility, cell surface proteins, immune clonotype, antigen specificity, and CRISPR perturbation screens.
Features
-
Small benchtop instrument (29x48x27cm)
-
Touch screen interface and retractable temperature controlled drawer
-
Fast workflow from cell suspension to sequencing-ready library
-
Captures 100-10,000 cells or nuclei per sample and up to 80,000 cells/nuclei per run
-
Variety of protocols available including analysis of fresh, frozen, or fixed samples for human or mouse
-
Compatible with Illumina sequencers
Other Instruments
Library preparation & preparation for sequencing
Quality control: Agilent 2100 BioAnalyzer, Agilent TapeStation 4150
Fluorometer: Invitrogen Qubit 4
Spectrophotometer: NanoDrop 1000

